TYPE: Research Article![]()
Preliminary Evidence of Hybridisation Between Golden Jackals and Dogs in the Malabar Region of Kerala, India: Insights from Mitochondrial DNA and Morphology
Kullumpurath P. Nidhichand¹, Muhasin Asaf²*![]() , George Chandy¹, Bindya Liz Abraham², K. Venkat Meganath², Ramesh Roshnath¹, Vijayakumar Sreeshma¹, Ivica Bošković³
, George Chandy¹, Bindya Liz Abraham², K. Venkat Meganath², Ramesh Roshnath¹, Vijayakumar Sreeshma¹, Ivica Bošković³
¹Centre for Wildlife Studies, Kerala Veterinary and Animal Sciences University, Pookode, India
²Department of Animal Genetics and Breeding, College of Veterinary and Animal Sciences, Pookode, Kerala Veterinary and Animal Sciences University, India
³Faculty of Agrobiotechnical Sciences Osijek, Department of Animal Production & Biotechnology, Chair for Hunting and Fishery, Vladimira Preloga 1, 31000 Osijek, Croatia
RECEIVED 20 August 2025
ACCEPTED 24 November 2025
ONLINE EARLY 23 December 2025
Abstract
Hybridisation between wild and domestic canids presents an increasing conservation concern, particularly in areas where human activities facilitate contact between species. This study investigates hybridisation between golden jackals (Canis aureus) and dogs (Canis lupus familiaris) in the Malabar region of Kerala, India. Suspected hybrids were initially identified through intermediate morphological traits, including coat colour, ear shape, and tail morphology. Mitochondrial DNA analysis of D-loop sequences from blood, stool, and tissue samples further supported these findings. Phylogenetic reconstruction using the maximum likelihood and Bayesian methods revealed that putative hybrids clustered with golden jackals in a distinct clade. A median-joining haplotype network showed shared haplotypes between hybrids and jackals, confirming maternal lineage from jackals. The presence of hybrids raises critical conservation concerns, potentially threatening the genetic integrity and ecological roles of jackals. In the future, continued genetic monitoring combined with the inclusion of additional nuclear markers will be essential for understanding hybridization dynamics and informing effective conservation and management strategies.
Keywords: Canid hybrids, dogs, golden jackal, hybridisation, mitochondrial DNA, phylogenetics
Introduction
Human–wildlife interactions are increasing globally due to expanding human populations and habitat encroachment, often leading to heightened interactions between wild and domesticated animals (Hindrikson et al., 2012). Among these, free-ranging dogs (Canis lupus familiaris) pose a significant ecological threat through direct predation, competition, and scavenging (Vanak & Gompper, 2009). Additionally, stray dogs are known vectors of several zoonotic and epizootic diseases, including rabies (Hueffer & Murphy, 2018), canine distemper (Deem et al., 2000), and canine parvovirus (Nandi & Kumar, 2010). Beyond these direct impacts, hybridisation between domestic dogs and wild canids presents a critical conservation concern, as it can lead to genetic introgression, reduced fitness, and loss of unique evolutionary lineages (Randi, 2008; Wegner & Eizaguirre, 2012).
The Indian or golden jackal (Canis aureus), is a small, canid distributed widely across the Indian subcontinent. It exhibits dusky yellowish or rufous-grey fur mottled with black, grey, and brown hairs, brownish-yellow underfur, a reddish-brown tail with a dark tuft, and rufous-coloured limbs and muzzle (Vattakaven et al., 2016). The Indian Pariah dog (Canis lupus familiaris), also known as the Desi Dog, INDog, or Pye-Dog, is an ancient breed native to the region. These dogs are distinguished by their erect ears, wedge-shaped head, curled tail, and short, variably coloured coat (Samir & Alsad, 2023).
Hybridisation between wild canids and dogs has been well documented in different regions. Cases include crosses between dogs and Ethiopian wolves (Canis simensis) in the Ethiopian highlands (Gottelli et al., 1994), grey wolves in the Indian savannahs (Tyagi et al., 2023), and Italian wolves in Europe (Randi et al., 2000; Randi, 2008). Similar hybridisation events have been recorded in Croatia (Kusak et al., 2018), Estonia and Latvia (Hindrikson et al., 2012), and even between Pampas foxes and dogs in South America (Szynwelski et al., 2023). Golden jackal–dog hybrids have recently been confirmed in Hungary’s Carpathian Basin (Ninausz et al., 2023) and in Croatia (Galov et al., 2015), yet no such hybrids have been scientifically documented in India to date.
Although unverified sightings and anecdotal accounts suggest the presence of jackal–dog hybrids in India, there remains a lack of scientific validation. Given the evolutionary proximity between jackals and dogs, the potential for hybridisation is plausible and warrants formal investigation (Shameer et al., 2022). This study was therefore undertaken to identify and characterise putative golden jackal–dog hybrids along the Malabar Coast of Kerala, India.
Materials and Methods
The study was approved by the Kerala Forests and Wildlife Department, Government of Kerala as per the order KFDHQ/1261/2024-CWW/WL10 dated 19.03.2024 of the Principal Chief Conservator of Forests (Wildlife) and Chief Wildlife Warden, Kerala. The study locations included the districts of Kannur, Kozhikode, and Malappuram in Kerala, along the Malabar Coast of southern India. This region, situated between the Arabian Sea and the Western Ghats, features a humid tropical monsoon climate, characterised by long rainy seasons, hot to warm summers, and mild, short winters. The area’s diverse ecosystems including coastal plains, agricultural landscapes, and fragmented forest patches provided an ideal setting for investigating golden jackals (Canis aureus), free-ranging dogs (Canis lupus familiaris), and their potential hybrids. The primary aim was to identify and map suspected jackal–dog hybrid zones using a citizen science approach.
Observational sampling was employed during peak activity periods: early morning (05:00–10:00), midday (11:00–14:00), and late evening (15:00–20:00), across the years 2022 to 2024. Field surveys focused on documenting animal morphology, resting sites, suspected hybrid individuals and pack size.
The locations of jackal packs were first identified through systematic field surveys. Suspected hybrids were then identified based on distinct morphological characteristics such as coat colour, tail length morphology and ear shape. Data were collected from citizen scientists, including local photographers, birdwatchers, and members of nearby communities. Awareness posters featuring images of suspected hybrid individuals were distributed to aid in identification and encourage public engagement. This effort resulted in the identification of 18 locations in the study area. Each site was validated through photographic submissions, phone interviews, site visits, and extended field observations. A detailed map of these sampling sites was created using QGIS version 3.38.1. The sampling locations are illustrated in Figure 1.
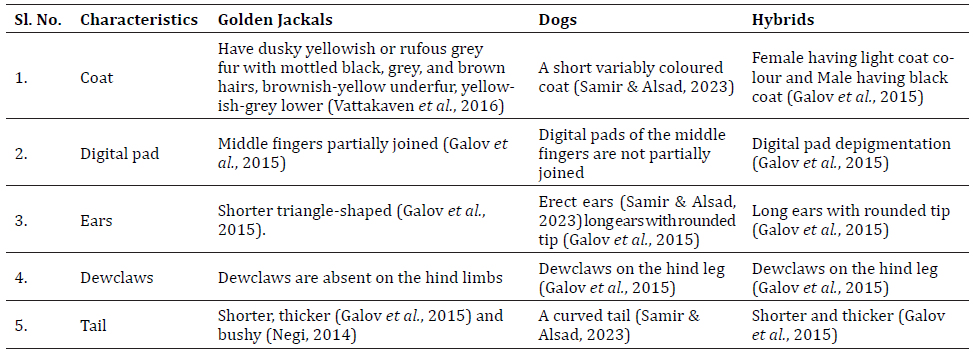
The morphological identification of putative golden jackal–dog hybrids was carried out based on diagnostic features reported in previous studies (Table 1). For the identification of putative hybrids based on morphology, we considered external characteristics such as coat colour, digital pads, ears, dewclaws, and tails. For body structure, the traits like robust body, coat colour, ear shape (pointed or rounded) and bushy tail were considered.
For the molecular genetic analysis, eight samples were utilised, comprising three golden jackals, three putative hybrids, and two dogs. Blood samples from dogs were collected during routine health examinations, and genomic DNA was extracted using the Wizard® Genomic DNA Purification Kit (Cat# A1120, Promega, USA). Faecal samples from suspected hybrids were preserved
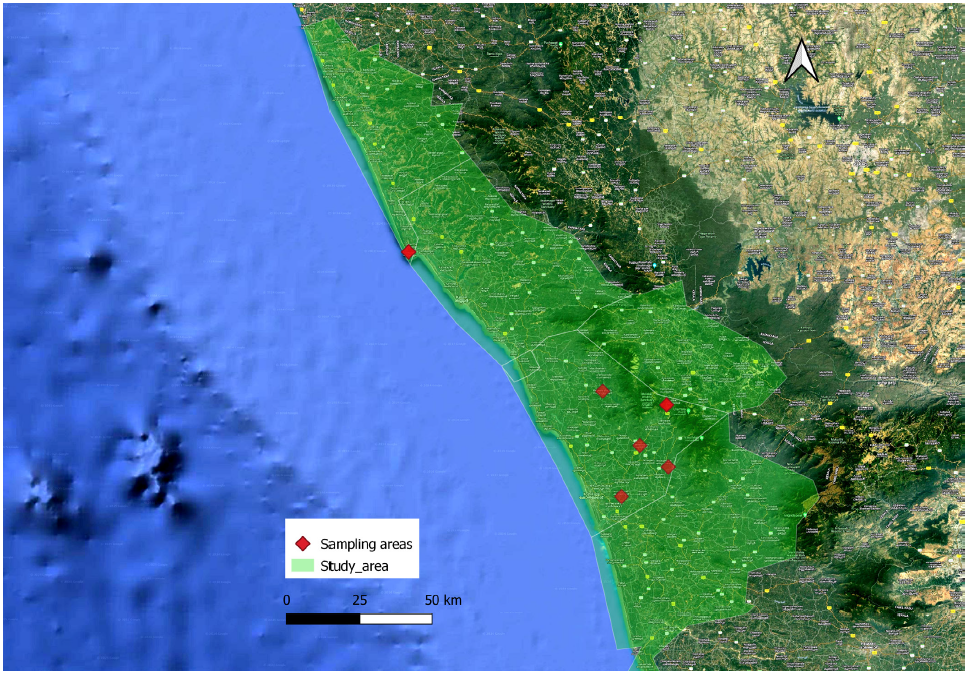
Figure 1: Study area representing the site locations of the genetic samples collected for the study.
Table 1. Morphological comparison of golden jackals, dogs, and putative hybrids based on external characteristics used in this study for hybrid identification.
in Longmire buffer (Longmire et al., 1997) and stored at room temperature until further processing. DNA from these samples was extracted using the QIAamp Fast DNA Stool Mini Kit (Cat# 51604, Qiagen, India). Muscle tissue samples from golden jackals were collected during postmortem examinations, and DNA was extracted using DNAiso Reagent (Cat# 9770A, DSS Takara Bio, India). All extractions were carried out according to the respective manufacturers’ protocols.
The concentration and purity of the extracted genomic DNA were assessed using a NanoDrop 2000C spectrophotometer (Thermo Fisher Scientific, USA). A 550 bp fragment of the mitochondrial hypervariable control region was amplified by PCR using primers L-Pro:5′-CGTCAGTCTCACCATCAACCCCCAAAGC-3′ and H576: 5′- TTTGACTGCATTAGGGCCGCGACGG-3′, as previously described (Douzery and Randi, 1997; Randi et al., 2000; Galov et al., 2015). PCR reactions were conducted in a 25 μL volume using EmeraldAmp GT Master Mix (Cat# RR310A, DSS Takara Bio, India). The thermal profile included an initial denaturation at 95 °C for 15 minutes, followed by 35 cycles of denaturation at 94 °C for 40 seconds, annealing at 60 °C for 50 seconds, and extension at 72 °C for 1 minute, with a final extension at 72 °C for 10 minutes. The amplified PCR products were visualiszed on a 1.8% agarose gel under a model UV transilluminator (Syngene Inc., USA).
Bands were purified using the GenElute™ Gel Extraction Kit (Cat# NA1111, Sigma-Aldrich, USA). Purified amplicons were sequenced using an automated Sanger dideoxy sequencing system. Resulting chromatograms were visualised and analysed using FinchTV v1.4.0 (https://digitalworldbiology.com/FinchTV). Final sequences were submitted to GenBank.
Multiple sequence alignment was performed using MAFFT v7 (Katoh & Standley, 2013), employing the L-INS-i algorithm for enhanced accuracy. Phylogenetic relationships were inferred using the maximum likelihood (ML) method implemented in IQ-TREE 2 (Minh et al., 2020) and Bayesian inference method in MrBayes 3.2.7 (Huelsenbeck et al., 2001). A median-joining haplotype network was constructed using PopART v1.7 (Leigh & Bryant, 2015).
Results and Discussion
Morphological analysis provided compelling evidence of hybridisation between golden jackals (Canis aureus) and dogs (Canis lupus familiaris). The suspected hybrids exhibited intermediate physical traits, particularly in coat colour, ear shape, and tail morphology, which are indicative of genetic mixing between jackals and dogs (Galov et al., 2015). These observations are consistent with reports from Hungary and Croatia, where morphological markers such as coat colour and ear morphology have been reliable indicators of hybridisation (Ninausz et al., 2023; Szynwelski et al., 2023).
Notably, this study documented a wide variety of coat colouration among hybrids, ranging from light brown and dark brown to blackish tones which is atypical for pure golden jackals (Figure 3). Some individuals displayed mixed patterns, including brownish fur with or without black patches on the tail tip. Black-coated individuals were particularly common at certain sites, marking a distinct deviation from typical jackal colouration. Other anomalies such as leucism, albinism, and rusty coats observed in isolated populations further support the potential influence of genetic mixing, although environmental factors may also play a role (Shameer et al., 2022; Sanil et al., 2014).
The ear shape of golden jackals is typically pointed, while their tails are bushy with a black tip. Among the hybrids, considerable variability was noted, while some retained the pointed ear structure of jackals, others had rounded or broader ears resembling those of dogs. Tail morphology also varied, from completely black-tipped tails to those lacking black markings, and tails that were less bushy or more elongated than those of pure jackals. Some individuals showed tails with only three-quarters of the tip darkened. Most of the hybrids, with the exception of a single individual, exhibited a pointed snout characteristic of golden jackals. In terms of body structure, they consistently displayed intermediate builds, larger and more robust than jackals, yet smaller than most dogs. Overall, the morphological traits documented in this study closely resemble those described by Galov et al. (2015), further supporting the likelihood of a hybrid origin (Figure 2). Representative photographs of the putative hybrids are presented in Figure 3.
The phylogenetic analysis was done using maximum likelihood (ML) and Bayesian methods. For this along with sequences generated from the current study, mitochondrial D-loop sequences retrieved from NCBI database were also utilised. Phylogenetic analysis of mitochondrial D-loop sequences using the ML method revealed distinct clades corresponding to golden jackals, wolves (Canis lupus), and dogs (Canis lupus familiaris), with the red fox (Vulpes vulpes) serving as the outgroup (Figure 4). The Indian wolf (AY289974.1) and Himalayan wolf (AY289995.1) were grouped as sister taxa to dogs, reflecting their shared evolutionary lineage. Golden jackals clustered with the hybrids, forming a distinct clade with strong bootstrap support (99%), where all reference and newly

Figure 2. Comparative photographs of jackal–dog hybrids. Top: Black-coated and brown-coated hybrids reported by Galov et al. (2015). Bottom: Corresponding black-coated and brown-coated hybrids documented in the present study

Figure 3. Photographic documentation of putative jackal–dog hybrids observed in the field. (A) Interaction between a dark brown-coated hybrid and a golden jackal. (B) Interaction between black-coated hybrids and golden jackals. (C) Black-coated hybrid sub-adult. (D) Light-coated hybrid sub-adult.
sequenced specimens grouped together. The red fox (AF338789.2) rooted outside all Canis lineages, supporting its use as an appropriate outgroup (Figure 4). The Bayesian phylogeny generated in MrBayes 3.2.7 resolved distinct clades for dogs and jackals with all hybrid individuals clustering with the jackal clade with the posterior probability values providing robust support for the major clades (Figure 5). The results were in agreement with the ML phylogenetic analysis.
A median-joining haplotype network generated using PopART v1.7 demonstrated that hybrid sequences shared haplotypes with golden jackals (Figure 6). The haplotype network showed distinct clusters corresponding to dog (D) and jackal (J) haplotypes. Hybrid (H) samples were distributed between these clusters. The results suggest that female jackals mated with male dogs, a pattern consistent with other canid hybridisation studies (Galov et al., 2015; Stronen et al., 2012), where maternal inheritance is typically linked to wild species and paternal inheritance to domestic ones (Gottelli et al., 1994; Lucchini et al., 2002). However, this study did not analyse nuclear DNA, leaving the paternal lineage and the full extent of genetic mixing unresolved. Future studies should incorporate nuclear markers to better assess hybridisation dynamics, as recommended in studies of Italian wolves and Ethiopian wolves (Randi et al., 2000; Freedman et al., 2014).
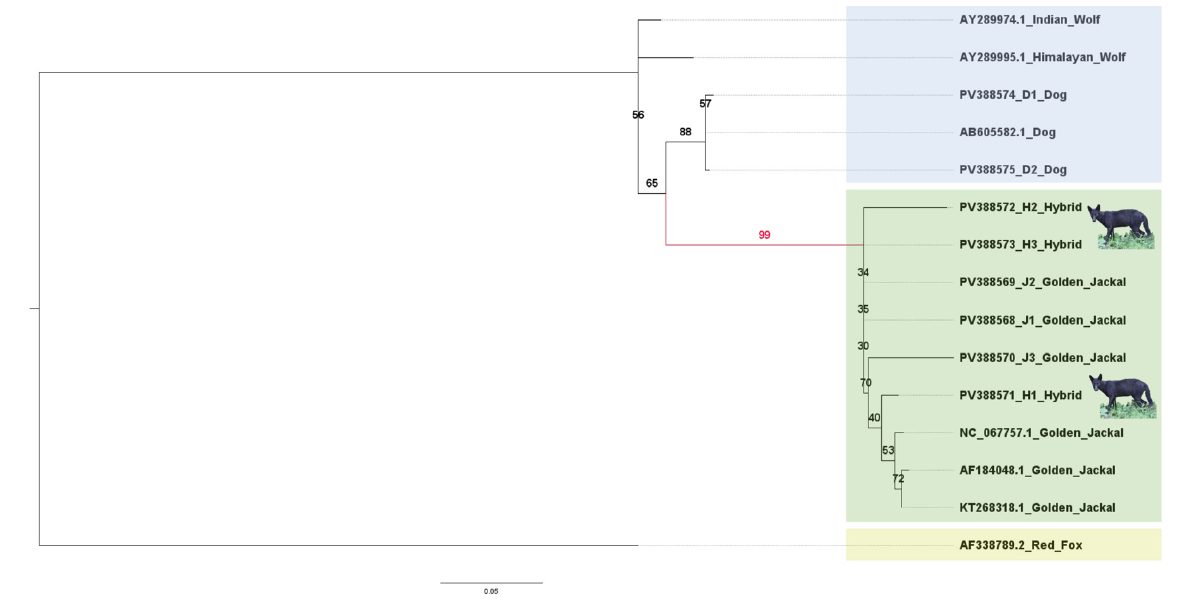
Figure 4: Maximum likelihood (ML) phylogenetic tree based on mitochondrial D-loop sequences showing the relationships among golden jackals (Canis aureus), dogs (Canis familiaris), putative hybrids, and related canid species. The tree was rooted using the red fox (Vulpes vulpes) as an outgroup. Bootstrap support values (based on 5000 replicates) are indicated at the nodes. Golden jackals and putative hybrids clustered together in a distinct clade with strong bootstrap support (99%), suggesting maternal lineage from jackals. Indian and Himalayan wolves grouped with dogs, consistent with their evolutionary proximity.
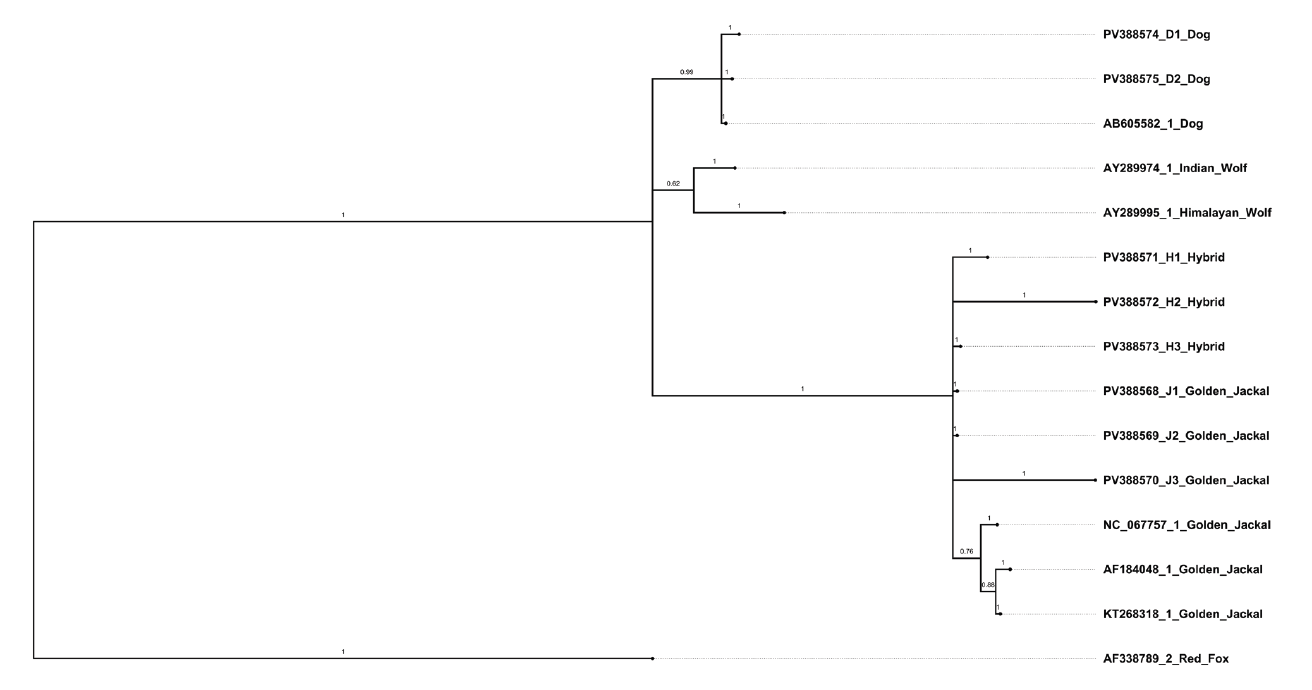
Figure 5: Phylogenetic tree generated by Bayesian inference method. The Bayesian phylogeny generated in MrBayes 3.2.7 resolved distinct clades for dogs and jackals. All hybrid individuals clustered with the jackal clade with strong posterior probability values provided robust support for the major clades.
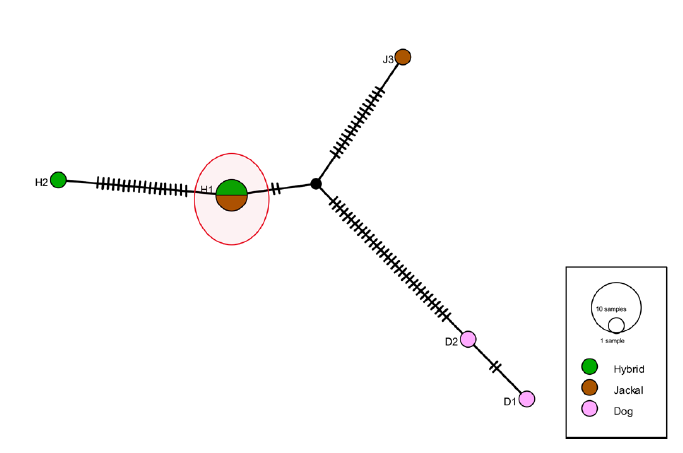
Figure 6: Median-joining haplotype network constructed using mitochondrial D-loop sequences from golden jackals (Canis aureus), dogs (Canis lupus familiaris), and putative hybrids. Each circle represents a unique haplotype, and the size of the circle is proportional to the number of individuals sharing that haplotype. The encircled haplotypes are shared between hybrids and golden jackals.
From a conservation perspective, the presence of golden jackal–dog hybrids poses significant ecological and management challenges. Hybridisation with dogs may dilute the genetic integrity of jackal populations, potentially reducing their adaptability and resilience (Randi, 2008; Wegner & Eizaguirre, 2012). To mitigate these risks, conservation strategies should prioritise the protection of golden jackal genetic integrity by monitoring hybrid occurrences and managing the effects of hybridisation. Recommended actions include sterilisation of free-ranging dogs near protected areas, public awareness campaigns on jackal conservation, and the maintenance of habitat corridors to reduce hybridisation risks in vulnerable zones.
This study underscores the importance of continued research into jackal – dog hybridisation. While our findings provide preliminary evidence for the presence of hybrids between jackals and dogs in Kerala, in future long-term monitoring and more comprehensive genetic studies including nuclear DNA are essential for fully understanding the extent, directionality, and ecological consequences of hybridisation. Conservation efforts should focus on preserving the genetic diversity of golden jackal populations along with addressing the broader ecological impacts of hybridisation through public engagement, policy development, and strategic habitat management. The current study has a limitation of a smaller sample size, absence of nuclear gene markers like microsatellites, SNPs. Hence, a larger study incorporating a larger sample size and genotyping using microsatellites and, SNP markers would provide a robust and deeper insight into the jackal-dog hybridisation.
TO DOWNLOAD SUPPLEMENTARY MATERIAL CLICK HERE.
Acknowledgement
We thank the Kerala Forests and Wildlife Department, Government of Kerala, for providing the necessary permissions for sample collection. We are also grateful to Subash Thomas and Tijo K. Joy at the KVASU–Centre for Wildlife Studies (KVASU-CWS) for their valuable assistance during lab work and fieldwork, respectively.
ETHICS STATEMENT
The study was approved by the Kerala Forests and Wildlife Department, Government of Kerala as per the order KFDHQ/1261/2024-CWW/WL10 dated 19.03.2024 of Principal Chief Conservator of Forests (Wildlife) and Chief Wildlife Warden, Kerala, and it involved no experiment or ethical issues.
CONFLICT OF INTEREST
The authors declare no competing interests.
DATA AVAILABILITY
Data and codes are available from the corresponding author on request.
AUTHORS’ CONTRIBUTION
GC and MA conceptualized the study. The methodology was developed by GC, BLA, and MA. Field investigation and data collection were carried out by NKP, RR, and SV. NKP and MA curated the data. Formal data analysis was performed by NKP, and MA. NKP prepared the original draft of the manuscript, while MA contributed to review and editing. MA supervised the study and was also responsible for data visualization. MKV, IB and MA provided resources for the study. Project administration was handled by MA.
USE OF GENERATIVE AI AND AI-ASSISTED TECHNOLOGIES
The author(s) used ChatGPT (Model: GPT-4) for language editing in the preparation of this manuscript. The author(s) thoroughly reviewed and edited the content generated and take(s) full responsibility for the content of the publication.
Edited By
Shrushti Modi
Wildlife Institute of India, Dehradun, India.
*CORRESPONDENCE
Muhasin Asaf
✉ muhasin@kvasu.ac.in
SPECIAL ISSUE
This paper is published in the Special Issue ‘Master’s Dissertations’. It was M.S. dissertation work of K.P. Nidhichand.
CITATION
Nidhichand, K. P., Asaf, M., Chandy, G., Abraham, B. L., Meganath, K. V., Roshnath, R., Sreeshma, V. & Bošković, I. (2025). Preliminary Evidence of Hybridisation Between Golden Jackals and Dogs in the Malabar Region of Kerala, India: Insights from Mitochondrial DNA and Morphology. Journal of Wildlife Science, Online Early Publication, 01-07. https://doi.org/10.63033/JWLS.OCBE3270
FUNDING
This research was supported by the KVASU– Centre for Wildlife Studies under the postgraduate research grant awarded to the first author (NKP).
COPYRIGHT
© 2025 Nidhichand, Asaf, Chandy, Abraham, Meganath, Roshnath, Sreeshma & Boskovic.This is an open-access article, immediately and freely available to read, download, and share. The information contained in this article is distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), allowing for unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited in accordance with accepted academic practice. Copyright is retained by the author(s).
PUBLISHED BY
Wildlife Institute of India, Dehradun, 248 001 INDIA
PUBLISHER'S NOTE
The Publisher, Journal of Wildlife Science or Editors cannot be held responsible for any errors or consequences arising from the use of the information contained in this article. All claims expressed in this article are solely those of the author(s) and do not necessarily represent those of their affiliated organisations or those of the publisher, the editors and the reviewers. Any product that may be evaluated or used in this article or claim made by its manufacturer is not guaranteed or endorsed by the publisher.
Deem, S. L., Spelman, L. H., Yates, R. A. & Montali, R. J. (2000). Canine distemper in terrestrial carnivores: a review. Journal of Zoo and Wildlife Medicine, 31(4), 441–451.
Douzery, E. & Randi, E. (1997). The mitochondrial control region of Cervidae: evolutionary patterns and phylogenetic content. Molecular Biology and Evolution, 14(11), 1154–1166.
Freedman, A. H., Gronau, I., Schweizer, R. M., Ortega-Del Vecchyo, D., Han, E., Silva, P. M., Galaverni, M., Fan, Z., Marx, P. & Lorente-Galdos, B. (2014). Genome sequencing highlights the dynamic early history of dogs. PLoS Genetics, 10(1), e1004016. https://doi.org/10.1371/journal.pgen.1004016
Galov, A., Fabbri, E., Caniglia, R., Arbanasić, H., Lapalombella, S., Florijanić, T., Bošković, I., Galaverni, M. & Randi, E. (2015). First evidence of hybridization between golden jackal (Canis aureus) and domestic dog (Canis familiaris) as revealed by genetic markers. Royal Society Open Science, 2, 150450. https://doi.org/10.1098/rsos.150450
Gottelli, D., Sillero-Zubiri, C., Applebaum, G. D., Roy, M.S., Girman, D. J., Garcia-Moreno, J., Ostrander, E. A. & Wayne, R. K. (1994). Molecular genetics of the most endangered canid: the Ethiopian wolf (Canis simensis). Molecular Ecology, 3, 301–312. https://doi.org/10.1111/j.1365-294x.1994.tb00070.x
Hindrikson, M., Männil, P., Ozolins, J., Krzywinski, A. & Saarma, U. (2012). Bucking the trend in wolf-dog hybridization: first evidence from Europe of hybridization between female dogs and male wolves. PLoS ONE, 7(10), e46465. https://doi.org/10.1371/journal.pone.0046465
Hueffer, K. & Murphy, M. (2018). Rabies in Alaska, from the past to an uncertain future. International Journal of Circumpolar Health, 77(1), 1475185. https://doi.org/10.1080/22423982.2018.1475185
Huelsenbeck, J. P. & Ronquist, F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics, 17(8), 754–755. https://doi.org/10.1093/Bioinformatics/17.8.754
Katoh, K. & Standley, D. M. (2013). MAFFT Multiple Sequence Alignment Software Version 7: Improvements in performance and usability. Molecular Biology and Evolution, 30(4), 772–780. https://doi.org/10.1093/molbev/mst010
Kusak, J., Fabbri, E., Galov, A., Gomerčić, T., Arbanasić, H., Caniglia, R., Galaverni, M., Reljić, S., Huber, D. & Randi, E. (2018). Wolf-dog hybridization in Croatia. Veterinarski Arhiv, 88, 375–395. https://doi.org/10.24099/vet.arhiv.170314
Leigh, J.W. & Bryant, D. (2015). PopART: full-feature software for haplotype network construction. Methods in Ecology and Evolution, 6(9), 1110–1116. https://doi.org/10.1111/2041-210X.12410
Longmire, J. L., Maltbie, M. & Baker, R. J. (1997). Use of “lysis buffer” in DNA isolation and its implication for museum collections. Museum of Texas Tech University, 163.
Lucchini, V., Fabbri, E., Marucco, F., Ricci, S., Boitani, L. & Randi, E. (2002). Non-invasive molecular tracking of colonising wolf (Canis lupus) packs in the western Italian Alps. Molecular Ecology, 11(5), 857–868. https://doi.org/10.1046/j.1365-294x.2002.01489.x.
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., von Haeseler, A. & Lanfear, R. (2020). IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Molecular Biology and Evolution, 37(5), 1530–1534. https://doi.org/10.1093/molbev/msaa015
Nandi, S. & Kumar, M. (2010). Canine parvovirus: current perspective. Indian Journal of Virology, 21(1), 31–44. https://doi.org/10.1007/s13337-010-0007-y
Negi, T. (2014). Review on current worldwide status, distribution, ecology and dietary habits of golden jackal, Canis aureus. Octa Journal of Environmental Research, 2(4), 338-359.
Ninausz, N., Fehér, P., Csányi, E., Heltai, M., Szabó, L., Barta, E., Kemenszky, P., Sándor, G., Janoska, F. & Horváth, M. (2023). White and other fur colourations and hybridisation in golden jackals (Canis aureus) in the Carpathian Basin. Scientific Reports, 13(1), 21969. https://doi.org/10.1038/s41598-023-49265-0
Randi, E., Lucchini, V., Christensen, M. F., Mucci, N., Funk, S. M., Dolf, G. & Loeschcke, V. (2000). Mitochondrial DNA variability in Italian and East European wolves: detecting the consequences of small population size and hybridisation. Conservation Biology, 14(2), 464–473. https://doi.org/10.1046/j.1523-1739.2000.98280.x
Randi, E. (2008). Detecting hybridisation between wild species and their domesticated relatives. Molecular Ecology, 17(1), 285–293. https://doi.org/10.1111/j.1365-294X.2007.03417.x
Samir, I. M. & Alsad, M. A. (2023). A brief review of the reproductive behaviour and behavioural diversity among Indian pariah dogs from Bangladesh. ESS Open Archive. https://doi.org/10.22541/essoar.169592585.54031335/v1
Sanil, R., Shameer, T. & Easa, P. S. (2014). Albinism in jungle cat and jackal along the coastline of the southern Western Ghats. CAT News, 61, 23–25.
Shameer, T. T., Bhavana, P. M., Mohan, G., Easa, P.S. & Sanil, R. (2022). Camera traps reveal coat colour variation in an isolated population of golden jackals. Current Science, 122(6), 738. https://doi.org/10.18520/cs/v122/i6/738-741
Stronen, A. V., Tessier, N., Jolicoeur, H., Paquet, P. C., Henault, M., Villemure, M., Patterson, 10 B. R., Sallows, T., Goulet, G. & Lapointe, F. J. (2012). Canid hybridisation: contemporary 11 evolution in human-modified landscapes. Ecology and Evolution, 2(9), 2128–2140. 12 https://doi.org/10.1002/ece3.335
Szynwelski, B. E., Kretschmer, R., Matzenbacher, C. A., Ferrari, F., Alievi, M. M. & Freitas, T. R. O. (2023). Hybridisation in canids—a case study of pampas fox (Lycalopex gymnocercus) and domestic dog (Canis lupus familiaris) hybrid. Animals, 13(15), 2505. https://doi.org/10.3390/ani13152505
Tyagi, A., Godbole, M., Vanak, A. T. & Ramakrishnan, U. (2023). Citizen science facilitates first ever genetic detection of wolf-dog hybridisation in Indian savannahs. Ecology and Evolution, 13, e10100. https://doi.org/10.1002/ece3.10100
Vanak, A. T. & Gompper, M. E. (2009). Dogs (Canis familiaris) as carnivores: their role and function in intraguild competition. Mammal Review, 39(4), 265–283. https://doi.org/10.1111/j.1365-2907.2009.00148.x
Vattakaven, T., George, R., Balasubramanian, D., Rejou-Méchain, M., Muthusankar, G., Ramesh, B. & Prabhakar, R. (2016). India Biodiversity Portal: an integrated, interactive and participatory biodiversity informatics platform. Biodiversity Data Journal, 4, e10279. https://doi.org/10.3897/BDJ.4.e10279
Wegner, K. M. & Eizaguirre, C. (2012). New(t)s and views from hybridising MHC genes: introgression rather than trans-species polymorphism may shape allelic repertoires. Molecular Ecology, 21(4), 779–781.
Edited By
Shrushti Modi
Wildlife Institute of India, Dehradun, India.
*CORRESPONDENCE
Muhasin Asaf
✉ muhasin@kvasu.ac.in
SPECIAL ISSUE
This paper is published in the Special Issue ‘Master’s Dissertations’. It was M.S. dissertation work of K.P. Nidhichand.
CITATION
Nidhichand, K. P., Asaf, M., Chandy, G., Abraham, B. L., Meganath, K. V., Roshnath, R., Sreeshma, V. & Bošković, I. (2025). Preliminary Evidence of Hybridisation Between Golden Jackals and Dogs in the Malabar Region of Kerala, India: Insights from Mitochondrial DNA and Morphology. Journal of Wildlife Science, Online Early Publication, 01-07. https://doi.org/10.63033/JWLS.OCBE3270
FUNDING
This research was supported by the KVASU– Centre for Wildlife Studies under the postgraduate research grant awarded to the first author (NKP).
COPYRIGHT
© 2025 Nidhichand, Asaf, Chandy, Abraham, Meganath, Roshnath, Sreeshma & Boskovic.This is an open-access article, immediately and freely available to read, download, and share. The information contained in this article is distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), allowing for unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited in accordance with accepted academic practice. Copyright is retained by the author(s).
PUBLISHED BY
Wildlife Institute of India, Dehradun, 248 001 INDIA
PUBLISHER'S NOTE
The Publisher, Journal of Wildlife Science or Editors cannot be held responsible for any errors or consequences arising from the use of the information contained in this article. All claims expressed in this article are solely those of the author(s) and do not necessarily represent those of their affiliated organisations or those of the publisher, the editors and the reviewers. Any product that may be evaluated or used in this article or claim made by its manufacturer is not guaranteed or endorsed by the publisher.
Deem, S. L., Spelman, L. H., Yates, R. A. & Montali, R. J. (2000). Canine distemper in terrestrial carnivores: a review. Journal of Zoo and Wildlife Medicine, 31(4), 441–451.
Douzery, E. & Randi, E. (1997). The mitochondrial control region of Cervidae: evolutionary patterns and phylogenetic content. Molecular Biology and Evolution, 14(11), 1154–1166.
Freedman, A. H., Gronau, I., Schweizer, R. M., Ortega-Del Vecchyo, D., Han, E., Silva, P. M., Galaverni, M., Fan, Z., Marx, P. & Lorente-Galdos, B. (2014). Genome sequencing highlights the dynamic early history of dogs. PLoS Genetics, 10(1), e1004016. https://doi.org/10.1371/journal.pgen.1004016
Galov, A., Fabbri, E., Caniglia, R., Arbanasić, H., Lapalombella, S., Florijanić, T., Bošković, I., Galaverni, M. & Randi, E. (2015). First evidence of hybridization between golden jackal (Canis aureus) and domestic dog (Canis familiaris) as revealed by genetic markers. Royal Society Open Science, 2, 150450. https://doi.org/10.1098/rsos.150450
Gottelli, D., Sillero-Zubiri, C., Applebaum, G. D., Roy, M.S., Girman, D. J., Garcia-Moreno, J., Ostrander, E. A. & Wayne, R. K. (1994). Molecular genetics of the most endangered canid: the Ethiopian wolf (Canis simensis). Molecular Ecology, 3, 301–312. https://doi.org/10.1111/j.1365-294x.1994.tb00070.x
Hindrikson, M., Männil, P., Ozolins, J., Krzywinski, A. & Saarma, U. (2012). Bucking the trend in wolf-dog hybridization: first evidence from Europe of hybridization between female dogs and male wolves. PLoS ONE, 7(10), e46465. https://doi.org/10.1371/journal.pone.0046465
Hueffer, K. & Murphy, M. (2018). Rabies in Alaska, from the past to an uncertain future. International Journal of Circumpolar Health, 77(1), 1475185. https://doi.org/10.1080/22423982.2018.1475185
Huelsenbeck, J. P. & Ronquist, F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics, 17(8), 754–755. https://doi.org/10.1093/Bioinformatics/17.8.754
Katoh, K. & Standley, D. M. (2013). MAFFT Multiple Sequence Alignment Software Version 7: Improvements in performance and usability. Molecular Biology and Evolution, 30(4), 772–780. https://doi.org/10.1093/molbev/mst010
Kusak, J., Fabbri, E., Galov, A., Gomerčić, T., Arbanasić, H., Caniglia, R., Galaverni, M., Reljić, S., Huber, D. & Randi, E. (2018). Wolf-dog hybridization in Croatia. Veterinarski Arhiv, 88, 375–395. https://doi.org/10.24099/vet.arhiv.170314
Leigh, J.W. & Bryant, D. (2015). PopART: full-feature software for haplotype network construction. Methods in Ecology and Evolution, 6(9), 1110–1116. https://doi.org/10.1111/2041-210X.12410
Longmire, J. L., Maltbie, M. & Baker, R. J. (1997). Use of “lysis buffer” in DNA isolation and its implication for museum collections. Museum of Texas Tech University, 163.
Lucchini, V., Fabbri, E., Marucco, F., Ricci, S., Boitani, L. & Randi, E. (2002). Non-invasive molecular tracking of colonising wolf (Canis lupus) packs in the western Italian Alps. Molecular Ecology, 11(5), 857–868. https://doi.org/10.1046/j.1365-294x.2002.01489.x.
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., von Haeseler, A. & Lanfear, R. (2020). IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Molecular Biology and Evolution, 37(5), 1530–1534. https://doi.org/10.1093/molbev/msaa015
Nandi, S. & Kumar, M. (2010). Canine parvovirus: current perspective. Indian Journal of Virology, 21(1), 31–44. https://doi.org/10.1007/s13337-010-0007-y
Negi, T. (2014). Review on current worldwide status, distribution, ecology and dietary habits of golden jackal, Canis aureus. Octa Journal of Environmental Research, 2(4), 338-359.
Ninausz, N., Fehér, P., Csányi, E., Heltai, M., Szabó, L., Barta, E., Kemenszky, P., Sándor, G., Janoska, F. & Horváth, M. (2023). White and other fur colourations and hybridisation in golden jackals (Canis aureus) in the Carpathian Basin. Scientific Reports, 13(1), 21969. https://doi.org/10.1038/s41598-023-49265-0
Randi, E., Lucchini, V., Christensen, M. F., Mucci, N., Funk, S. M., Dolf, G. & Loeschcke, V. (2000). Mitochondrial DNA variability in Italian and East European wolves: detecting the consequences of small population size and hybridisation. Conservation Biology, 14(2), 464–473. https://doi.org/10.1046/j.1523-1739.2000.98280.x
Randi, E. (2008). Detecting hybridisation between wild species and their domesticated relatives. Molecular Ecology, 17(1), 285–293. https://doi.org/10.1111/j.1365-294X.2007.03417.x
Samir, I. M. & Alsad, M. A. (2023). A brief review of the reproductive behaviour and behavioural diversity among Indian pariah dogs from Bangladesh. ESS Open Archive. https://doi.org/10.22541/essoar.169592585.54031335/v1
Sanil, R., Shameer, T. & Easa, P. S. (2014). Albinism in jungle cat and jackal along the coastline of the southern Western Ghats. CAT News, 61, 23–25.
Shameer, T. T., Bhavana, P. M., Mohan, G., Easa, P.S. & Sanil, R. (2022). Camera traps reveal coat colour variation in an isolated population of golden jackals. Current Science, 122(6), 738. https://doi.org/10.18520/cs/v122/i6/738-741
Stronen, A. V., Tessier, N., Jolicoeur, H., Paquet, P. C., Henault, M., Villemure, M., Patterson, 10 B. R., Sallows, T., Goulet, G. & Lapointe, F. J. (2012). Canid hybridisation: contemporary 11 evolution in human-modified landscapes. Ecology and Evolution, 2(9), 2128–2140. 12 https://doi.org/10.1002/ece3.335
Szynwelski, B. E., Kretschmer, R., Matzenbacher, C. A., Ferrari, F., Alievi, M. M. & Freitas, T. R. O. (2023). Hybridisation in canids—a case study of pampas fox (Lycalopex gymnocercus) and domestic dog (Canis lupus familiaris) hybrid. Animals, 13(15), 2505. https://doi.org/10.3390/ani13152505
Tyagi, A., Godbole, M., Vanak, A. T. & Ramakrishnan, U. (2023). Citizen science facilitates first ever genetic detection of wolf-dog hybridisation in Indian savannahs. Ecology and Evolution, 13, e10100. https://doi.org/10.1002/ece3.10100
Vanak, A. T. & Gompper, M. E. (2009). Dogs (Canis familiaris) as carnivores: their role and function in intraguild competition. Mammal Review, 39(4), 265–283. https://doi.org/10.1111/j.1365-2907.2009.00148.x
Vattakaven, T., George, R., Balasubramanian, D., Rejou-Méchain, M., Muthusankar, G., Ramesh, B. & Prabhakar, R. (2016). India Biodiversity Portal: an integrated, interactive and participatory biodiversity informatics platform. Biodiversity Data Journal, 4, e10279. https://doi.org/10.3897/BDJ.4.e10279
Wegner, K. M. & Eizaguirre, C. (2012). New(t)s and views from hybridising MHC genes: introgression rather than trans-species polymorphism may shape allelic repertoires. Molecular Ecology, 21(4), 779–781.




